About Duplicates
This week I got some questions about duplicates. In order to really know how to analyse your data, you must know the possible problems of duplicates. But, what are duplicates?
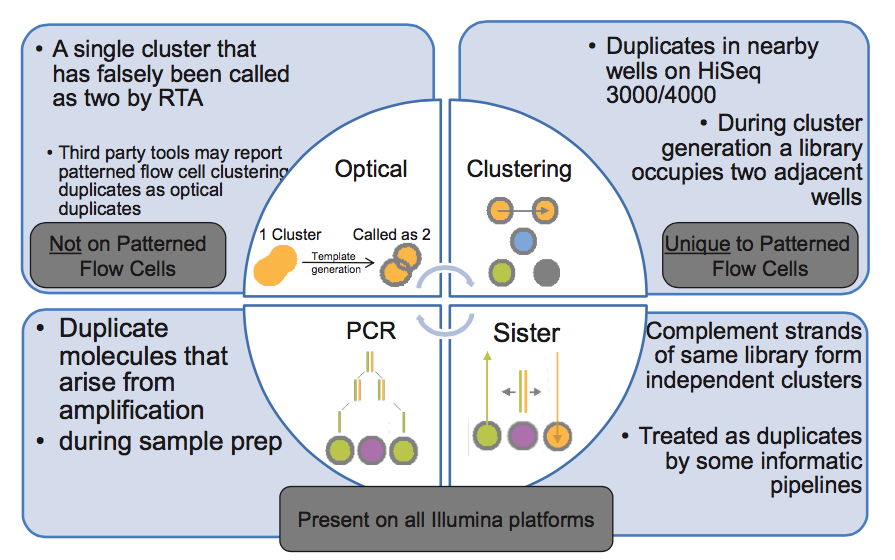
Actually there are different kinds of duplicates. The most logical kind are pcr duplicates. These are exact copies of the same DNA fragment. This means the fragment start and ends on the same location on the genome. Which makes it easy to recognize in your data. Several tools exist to mark or remove them (like Picard and elPrep). Removing them is an obvious thing to do, but there are some exceptions. Lab protocols using restriction enzymes (like GBS, RAD, …) or primers (amplicons) creates fragments that are starting and ending on the same position. Executing this duplicate removal tools on this data will resolve in only 1 read per location.
Another kind of duplicates are those introduced by the sequencing. In Illumina Sequencers fragments are clustered on flowcells. These flowcells are seperated in lanes. These lanes are the smallest units you can load with samples. However to be able to read these lanes in a high resolution, these lanes are split into tiles. Each corporation of a base, a picture of a tile is taken. In order not to miss clusters on tile borders, tiles are overlapping. This means that clusters in these overlaps will be seen twice. So this are optical duplicates. However, since they are on the same location, and have the same coordinate, filtering them out is pretty easy. However you do not need to worry about these, since the Illumina software removes them during demultiplexing.
The clusters on the flowcells are detected by the software, but sometimes these clusters are rather big, and are falsly split into 2 exactly the same clusters. These are optical duplicates. In the data they appear exactly the same as pcr duplicates, and can be removed the same way.
The latest Illumina sequencers contain the Illumina patterned flow cell. These flowcells have a system were the clusters are not random distributed on the flowcell, but are instead organized into wells. While generating the cluster inside a well, a fragment can go accedentally to a nearby well. This generates a new duplicate, very similar to the optical duplicates from the previous systems.
Overall duplicates are fairly easy to find. Several tools are available to mark or delete them (for example Picard and elPrep). Most important is to know the used lab protocol, in order to know if you need to remove them or not.
More info about duplicates can be found on this site